|
NMR Prediction: The handling of molecular structures within Mnova also allows the user to carry out spectral predictions. Although the process described here could be applied to predictions by means of any commercially available or in-house prediction package, currently Mnova will carry out predictions by using the prediction application NMRPredict, by Modgraph Consultants Ltd. Please note that this is a separate plugin which must be licensed in addition to Mnova.

|
A few notes on NMR Prediction Server-based
✓As opposed to NMRPredict Desktop, NMRPredict server-based is a client-server application, so to carry out the predictions you will have to log in to the server via a dialog box shown by Mnova. Log in details will be provided to you when downloading NMRPredict. This login will only have to be carried out the first time you run a prediction with Mnova. After that, the software should remember your login details and do the log in in the background. ✓Because it is a client-server application and the data are being sent over the internet to the server, industrial testers may wish to have an in-house installation of NMRPredict for security reasons. If this is the case, just write to us with your request at nmrpredict@mestrelab.com. ✓Customers who have their own in-house prediction package are welcome to contact us to discuss a similar, seamless integration of their own prediction package. Please write to us at support@mestrelab.com. ✓Please note that NMRPredict is a different software package to Mnova and therefore, in order to carry out predictions via this package, a license of NMRPredict must be held separately, once the NMRPredict evaluation has expired.
|
Carrying out of predictions in Mnova is very simple. With a molecular structure open in the active page of Mnova, just go to the 'Prediction' menu and select the applicable Prediction Options; then a dialog box will open (One for each Nuclei).
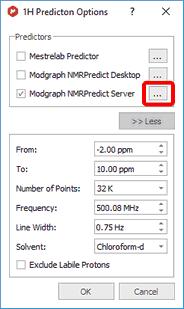
This open dialog allows the user to set a series of prediction parameters before carrying out the prediction: limits, number of points, frequency of the spectrometer, line width, solvent and the Predictor methodology. To carry out a server-based NMR prediction the user has to select 'Modgraph NMRPredict Server' in the 'Predictor' edit box.
Clicking on the '...' (red square in the capture above), the user will be able to select the algorithm for the 1H-NMR prediction (Increment or Charge):
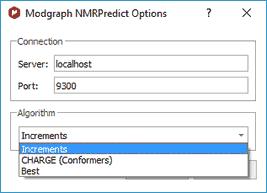
The Increment algorithm (based in Ernö Pretsch´s NMR tables) is the same as NMRPredict Desktop uses to predict NMR spectra
The CHARGE program offers 1H-NMR prediction based upon partial atomic charges and steric interactions. This program offers the first quantitative prediction of the proton chemical shifts of a variety of organic compounds.
The program works by first generating 3D conformers from a 2D structure through a choice of force fields and then predicting proton spectra for all the conformers. The program works by looking at functional groups which have parametrised by Professor Abraham.
To read more about the CHARGE theory, please visit the following link:
http://www.modgraph.co.uk/product_nmr_proton.htm

|
Please remember that:
NMRPredict is a client-server application, so to carry out the predictions you will have to log in to the server via a dialog box shown by Mnova. Log in details will be provided to you when downloading NMRPredict. This login will only have to be carried out the first time you run a prediction with Mnova. After that, the software should remember your login details and do the log in in the background.
|
Finally, just go to the 'Prediction' menu and select the desired prediction.
This will send a request to the server, which will run a prediction and return a set of values which Mnova will display as a spectrum on the screen. You will immediately notice that this will be an assigned spectrum, and therefore hovering of the mouse over the atoms will highlight the applicable peak on the spectrum and viceversa.

|
A few notes on the synthetization of NMR 1H spectra
✓It is important to note that the simulation carried out by Mnova when displaying the predicted values as a spectrum is not first order. The software is carrying out the full quantum-mechanics calculations to arrive at the displayed spectrum. This is very computationally intensive and results in such simulation module having a limitation, for 1H spectra, which prevents it from simulating spin systems larger than 13 spins. However, we have developed a new algorithm to overcome this problem.
|
The prediction of the 13C-NMR is based on the HOSE code approach. The HOSE (Hierarchical Organisation of Spherical Environments) code approach starts at the carbon atom whose shift is to be predicted, looks one bond away from the carbon and tries to find this same environment in its database. If it is successful it moves two bonds away and tries again and so on until it either comes across something not represented in the database or it reaches the boundary of the molecule.
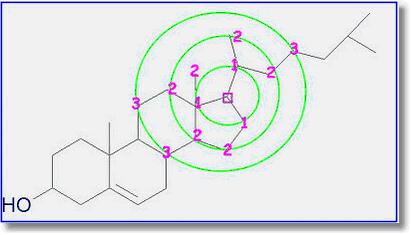
The HOSE code approach works very well for query structures which are well represented in the reference collection. If atoms can be predicted to three spheres or more (NMRPredict goes to a maximum of five spheres) the prediction can be considered to be very reliable.
However, if the query structure is not well represented in the database and the atom can only be predicted to one or two spheres the prediction cannot be relied upon at all. Also the HOSE code approach exactly reproduces the contents of the reference database – including every error within the reference database.
If atoms can only be predicted to one or two spheres the only solution traditionally was to add your own similar data and use that for prediction. NMRPredict also uses a unique Neural Network algorithm, which is much more general and error tolerant than the HOSE code approach and is much more accurate at predicting atoms it has not seen in its database.
| 




