|
Mnova incorporates a plugin to process LC/GC chromatograms and mass spectra. The concept behind the multiplatform, multipage Graphical User Interface of Mnova is to allow the average chemist easy access to high quality processing and analysis of analytical data on the desktop computer and OS of choice. The MSChrom plugin gives the chemist the ability to work with LC/GC/MS data in combination with NMR data, within a single document.
The Mnova GUI gives you much more. Benefit from day one of many other Mnova capabilities, such as scripting, reporting templates, graphical annotations, multiple exporting formats, including PDF, OLE integration with MS Office and many, many more.
In the picture below you can see an example of a layout template used for the MS plugin:
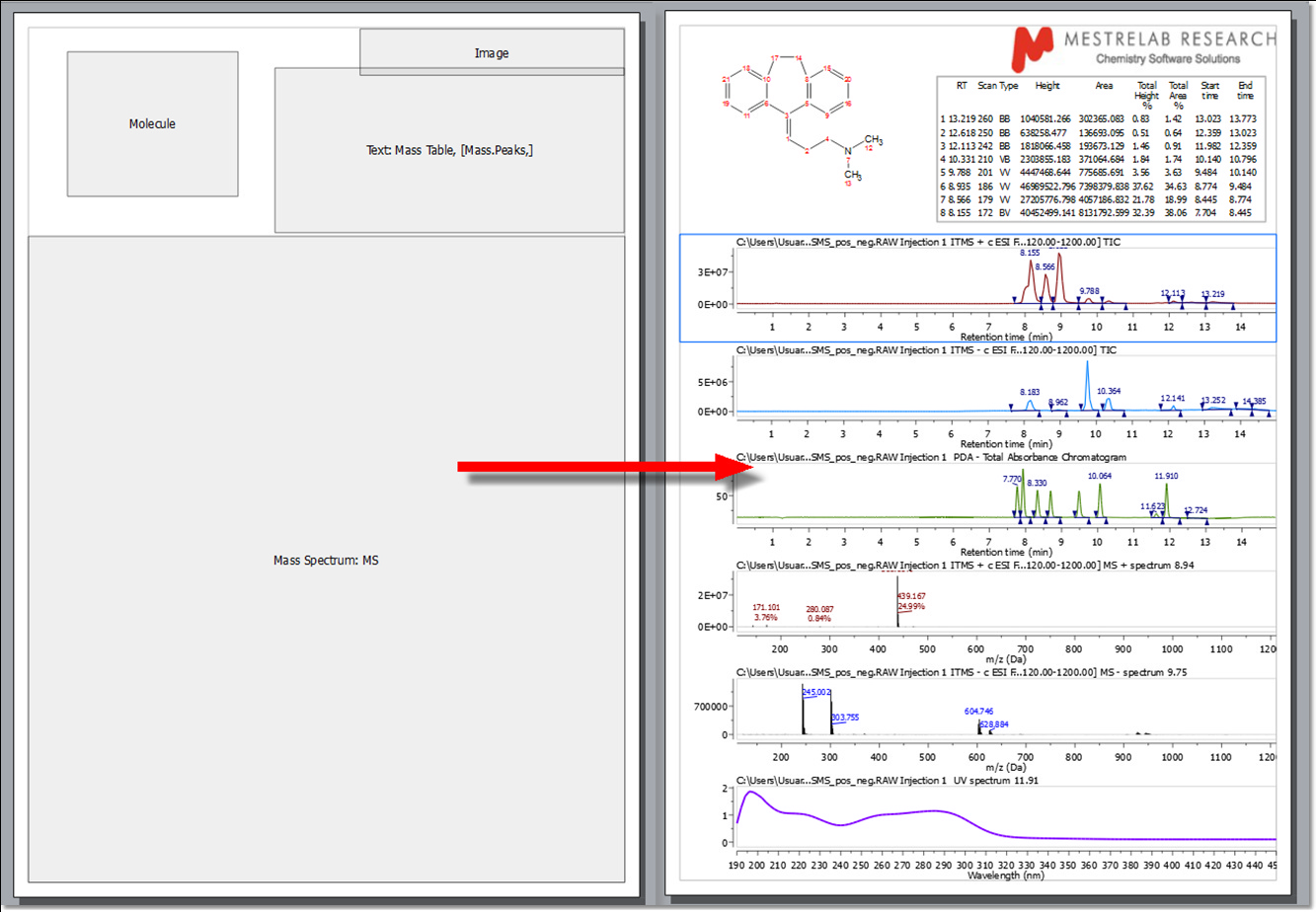
Mnova is an extremely versatile reporting tool, which will allow users to report even NMR and LC/GC/MS data together in the same document (or page):
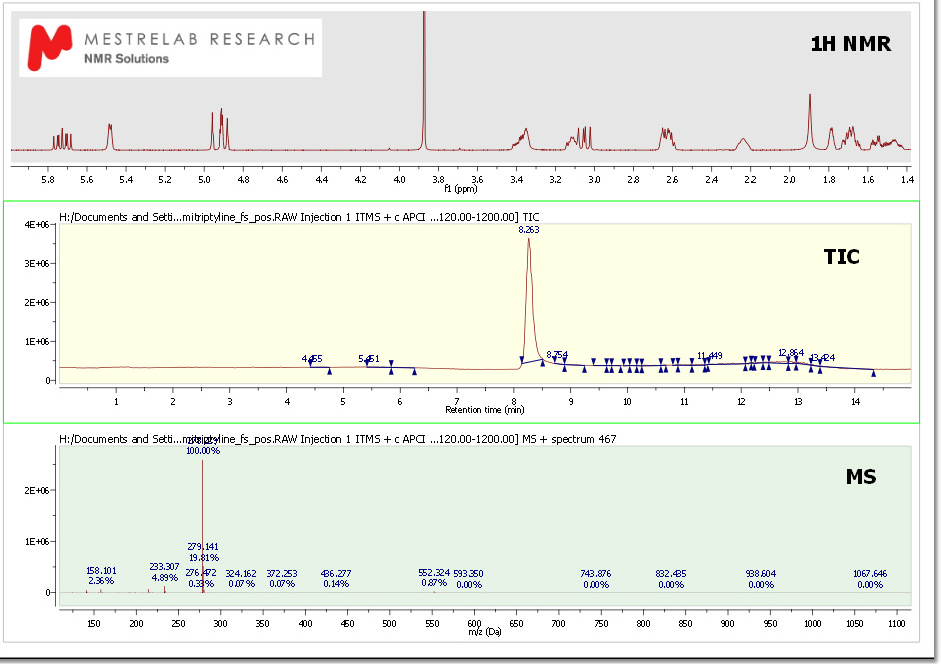
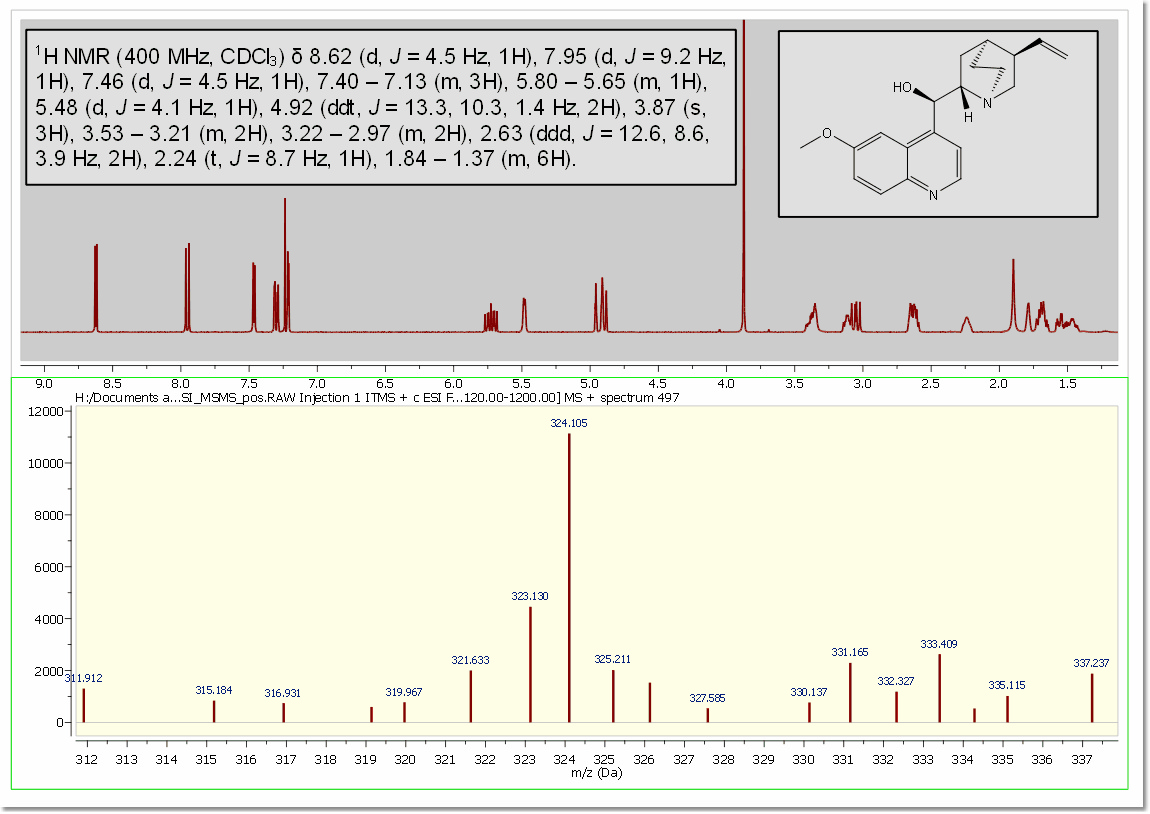
Here you can see another example with a MS and a 1H-NMR spectrum, containing a molecule structure and multiplet report:
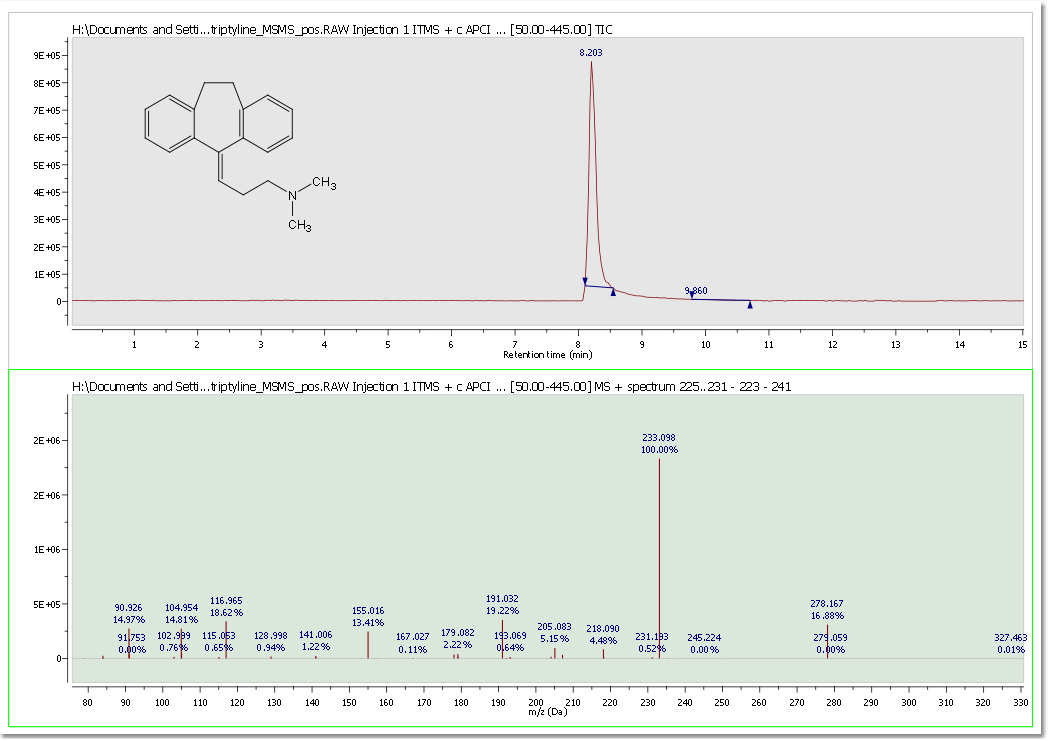
Following the Mnova concept, seamlessly interact with and handle data from different vendors. HP Agilent (ChemStation, Ion trap, MassHunter), Thermo Scientific–Xcalibur, Waters MassLynx, Waters UNIFI, Bruker (Compass, XMass), JEOL MSQ1000, Accu-ToF 4G LC-Plus, SCIEX Analyst, Sciex Data Explorer (after having run DE software with admin privileges, Shimadzu, NetCDF ANDI-MS, Varian Saturn, mzData. mzML and mzXML. It provides a graphical user interface (GUI) for interaction with and display of dataset contents and tools and algorithms for basic processing of chromatograms and mass spectra.
Please bear in mind that Mnova MSChrom under Mac and Linux only supports:
•Agilent – ChemStation, Ion Trap •Bruker Compass •JEOL – MSQ1000 •MS CSV File •Waters OpenLynx Report •Waters – MassLynx (Mac only) •mzData and mzXML Here you can find the list of supported MS formats by OS:
https://resources.mestrelab.com/ms-supported-formats/
The target user of the MSChrom plug-in is one whose goals are the elucidation and confirmation of small molecule structures.
The Mnova framework will allow the user to combine the results of multiple analyses into a consolidated report of results.
You will find below a glossary of the main terms relating to the MS plugin:
•Adduct: Ion formed by the interaction of an ion with one or more atoms or molecules to form an ion containing all the constituent atoms of the precursor ion as well as the additional atoms from the associated atoms or molecules. | • | Accurate Mass: Experimentally determined mass of an ion that is used to determine an elemental formula. Typically, a mass accuracy of 5 ppm or better, at relatively low m/z values, is necessary to compute a correct elemental formula. See the section on elemental composition later in this document. |
| • | Base Peak: The most intense peak of the mass spectrum which is assigned a relative abundance value of 100 percent. The lesser peaks are reported as a fraction of the base peak abundance. |
| • | Chemical ionization (CI): Formation of an ion in the gas phase by the reaction of a neutral species with an ion. The process may involve transfer of an electron, a proton or other charged species between the reactants. Note: When a positive ion results from chemical ionization the term may be used without qualification. When a negative ion results the term negative ion chemical ionization should be used. |
| • | Electron Ionization (EI): Ionization of an atom or molecule by electrons that are typically accelerated to energies between 10 and 150 eV in order to remove one or more electrons from the molecule. The term electron impact is deprecated. |
| • | Electrospray Ionization (ESI): A process in which ionized species in the gas phase are produced from a solution via highly charged fine droplets, by means of spraying the solution from a narrow-bore needle tip at atmospheric pressure in the presence of a high electric field (1,000 to 10,000 V potential). |
| • | Fast Atom Bombardment (FAB): A method in which ions are produced in a mass spectrometer from non-volatile or thermally fragile organic molecules by bombarding the compound in the condensed phase with energy-rich neutral particles. |
| • | Isotope Cluster: Set of mass peaks related to ions with the same chemical formula but containing different isotopes; e.g. the 16 and 17 Da peaks in a CH4 sample arising from 12CH4+ and 13CH4+ ions. |
| • | Mass Spectrometry/Mass Spectrometry (MS/MS): The acquisition and study of the spectra of the electrically charged products or precursors of m/z selected ion or ions, or of precursor ions of a selected neutral mass loss. MS/MS can be accomplished using beam instruments incorporating more than one analyzer (tandem mass spectrometry in space) or in trap instruments (tandem mass spectrometry in time). |
| • | Mass Spectrum: A plot of the relative abundances of ions as a function of their m/z (mass to charge ratio) values, representing data produced by a mass spectrometer. |
| • | Mass Resolution: In a mass spectrum, the smallest mass difference (Δm) between two equal magnitude mass peaks such that the valley between them is a specified fraction of the peak height. Typically, a 10% valley is specified, or 50%, which is known as full width at half-maximum height (FWHM). |
| • | Mass Resolving Power: In a mass spectrum, the observed mass divided by the difference between two masses that can be separated: m/Δm. |
| • | Mass Chromatogram: A chromatogram obtained from the plot of the intensity of a single m/z value or of a range of m/z values for each mass spectra in a dataset versus time. |
| • | Molecular Isotope Cluster Chromatogram (MICC): The MICC is the sum of the mass chromatograms of each m/z value of an isotope cluster. |
| • | Total Ion Chromatogram (TIC): The chromatogram obtained from the plot of the sum of intensities for each mass spectrum in a dataset versus time. |
|





